MSACL 2019 EUSalzburg Sept 22-26 |
Details
MSACL EU
|
Short Courses
MSACL hosts a diverse offering of Short Courses.
Short Courses will occur over the first three days of MSACL (Sunday, Monday, Tuesday of September 22 - 26).
Courses are NOT replicated on different days. They are single courses that span 1 or 2 days.
| Course Contact Hours | ||||||
| Sunday PM | Monday AM | Lunch | Monday PM | Tuesday AM | ||
| Data Science | ||||||
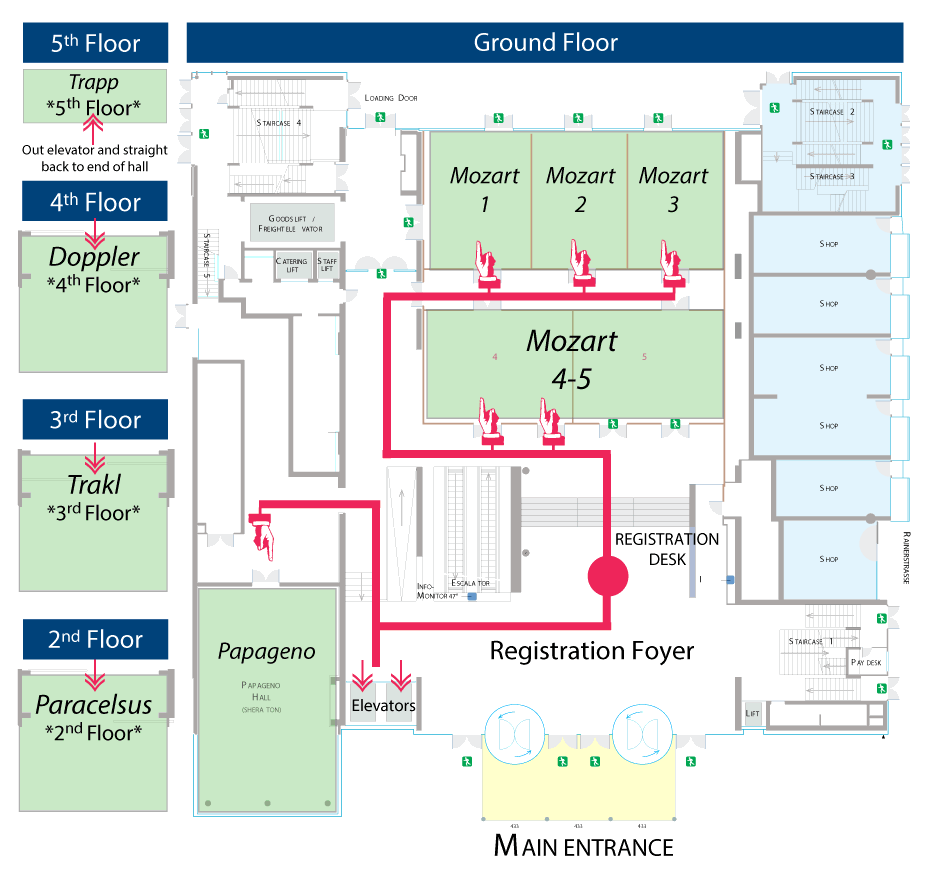
| Data Science 101 (Beginner - Intermediate) Breaking up with Excel: An Introduction to the R Statistical Programming Language Daniel Holmes, MD, William Slade, PhD & Stephen Master, MD, PhD Location: Trakl Hall | STARTS < Sunday 13:00-18:00 | Monday 8:30-12:30 | Lunch Monday 12:30-13:30 | Monday 13:30-17:30 | > ENDS Tuesday 8:30-11:30 | |
| Data Science 201 (Intermediate) Going Further With R: Tackling Clinical Laboratory Data Manipulation and Modeling Patrick Mathias, MD, PhD & Shannon Haymond, PhD Location: Mozart 3 | STARTS < Sunday 13:00-18:00 | Monday 8:30-12:30 | Lunch Monday 12:30-13:30 | Monday 13:30-17:30 | > ENDS Tuesday 8:30-11:30 | |
| Glyco-Proteomics | ||||||
| Glyco-Proteomics 101 (Beginner to Intermediate) Unravelling The Sweetness Of Life: Clinical Glyco(Proteo)Mics By Mass Spectrometry Noortje de Haan, PhD & Guinevere Lageveen-Kammeijer, PhD Location: Mozart 2 | STARTS < Sunday 13:00-18:00 | Monday 8:30-12:30 | Lunch Monday 12:30-13:30 | Monday 13:30-17:30 | > ENDS Tuesday 8:30-11:30 | |
| LC-MS | ||||||
| LC-MSMS 101 (Beginner) Getting Started with Quantitative LC-MS/MS in the Diagnostic Laboratory Laura Owen, PhD, Michael Wright, Coral Munday & Grace van der Gugten Location: Paracelsus Hall | STARTS < Sunday 13:00-18:00 | Monday 8:30-12:30 | Lunch Monday 12:30-13:30 | Monday 13:30-17:30 | > ENDS Tuesday 8:30-11:30 | |
| LC-MSMS 301 (Advanced) Development and Validation of Quantitative LC-MS/MS Assays for Use in Clinical Diagnostics Brian Rappold & Chris Shuford, PhD Location: Mozart 4-5 | STARTS < Sunday 13:00-18:00 | Monday 8:30-12:30 | Lunch Monday 12:30-13:30 | Monday 13:30-17:30 | > ENDS Tuesday 8:30-11:30 | |
| Lipidomics | ||||||
| Lipidomics 101 (Beginner - Intermediate) Mass Spectrometry-based Lipidomics and Clinical Applications Anne K Bendt, PhD, Amaury Cazenave-Gassiot, PhD, Michael Chen, MD Location: Papageno Hall | Not in Session | STARTS < Monday 8:30-12:30 | Lunch Monday 12:30-13:30 | > ENDS Monday 13:30-17:30 | Not in Session | |
| Metabolomics | ||||||
| Metabolomics 202 (Beginner to Intermediate) Metabolomics: Approaches, Applications and Challenges Julijana Ivanisevic, PhD & Elizabeth Want, PhD Location: Doppler Hall | STARTS < Sunday 13:00-18:00 | Monday 8:30-12:30 | Lunch Monday 12:30-13:30 | Monday 13:30-17:30 | > ENDS Tuesday 8:30-11:30 | |
| Microbiology | ||||||
| Proteomic Microbiology 201 (Beginner - Intermediate) Bottom-Up and Top-Down Proteomic Approaches for Bacterial Identification and Characterization, a Focus on MALDI-TOF and Advanced Technologies Jean Armengaud, PhD, Stefan Zimmermann, MD Location: Trapp Zimmer | Not in Session | Not in Session | Lunch Monday 12:30-13:30 | STARTS < Monday 13:30-17:30 | > ENDS Tuesday 8:30-11:30 | |
Notice: Undefined index: days in /home/msacl0017/public_html/include/webpage_structure/include_subtab_content_nextgen.php(1236) : eval()'d code on line 220
Data Science 101 :: Breaking up with Excel: An Introduction to the R Statistical Programming Language
| Level: | Beginner - Intermediate | ||||||||||||||||||||
| Prereqs: | Knowledge of Excel; able to bring a laptop; able to pre-install software on their laptop...Namely: R and R-studio; willingness to break up with Excel. | ||||||||||||||||||||
| Location: | Trakl Hall | ||||||||||||||||||||
| Instructor(s): | Daniel Holmes, MD, William Slade, PhD & Stephen Master, MD, PhD | ||||||||||||||||||||
| |||||||||||||||||||||
Daniel Holmes, MD did his undergraduate degree in Chemical Physics from the University of Toronto with a focus on Quantum Mechanics. He went to medical school at the University of British Columbia (UBC) where he also did his residency in Medical Biochemistry. He is a Clinical Associate Professor of Pathology and Laboratory Medicine at UBC and Division Head of Clinical Chemistry at St. Paul's Hospital in Vancouver. Interests include laboratory medicine statistics, clinical endocrinology, clinical lipidology and clinical mass spectrometry. Assay development efforts in the last two years have focused on assays specialized endocrine testing. William Slade, PhD did his undergraduate degree in Biology from Concord University with a focus on Recombinant Gene Technology. He completed a PhD in Biological Sciences at Virginia Tech with a focus on Biological Mass Spectrometry before postdoctoral work at the University of North Carolina at Chapel Hill in the Analytical Chemistry department. He is currently a Researcher in the Mass Spectrometry Research and Development group at Laboratory Corporation of America Holdings in Burlington, North Carolina. His interests include clinical mass spectrometry and informatics. Stephen Master, MD, PhD received his undergraduate degree in Molecular Biology from Princeton University, and subsequently obtained his MD and PhD from the University of Pennsylvania School of Medicine. After residency in Clinical Pathology at Penn, he stayed on as a faculty member with a research focus in mass spectrometry-based proteomics as well as extensive course development experience in bioinformatics. He is currently Associate Professor of Pathology and Laboratory Medicine at Weill Cornell Medicine in New York City, where he serves as Director of the Central Lab and Chief of Clinical Chemistry Laboratory Services. One of his current interests is in the applications of bioinformatics and machine learning for the development of clinical laboratory assays. He would play with R for fun even if he weren't getting paid, but he would appreciate it if you didn't tell that to his department chair. | |||||||||||||||||||||
| |||||||||||||||||||||
Notice: Undefined variable: late_reg in /home/msacl0017/public_html/include/webpage_structure/include_subtab_content_nextgen.php(1236) : eval()'d code on line 444
Overview: Have you ever tried to do Deming regression in Excel only to discover that it is not available? Have you had your figure rejected by a journal because the resolution was not good enough? Have you wished that you could figure out a way to stop manually transcribing your LC-MS/MS results into the LIS?
Well, your wait is over because this year at MSACL we will be offering a course for complete programming newbies that will help you get going analyzing real data related to LC-MS/MS assay development, validation, implementation and publication. The only background expected is the ability to use a spreadsheet program. The skills that you will acquire will allow you to take advantage of the many tools already available in the R language and thereafter, when you see that your spreadsheet program does not have the capabilities to do what you need, you will no longer have to burst into tears. You will be empowe-R-ed.
The course will be run over ~16 hrs and time will be evenly split between didactic sessions and hands-on problem solving with real data sets. The instructors will adopt a “no student left behind policy”. Students will be given ample time to solve mini problems taken from real life laboratory work and focused on common laboratory tasks. All attendees will need to bring a laptop with the R language installed R Studio interface installed. Students may use Windows, Mac OSX or Linux environments. Both R and R studio are free and open-source. No cash required.
Students should be prepared for learning what computer programming is really like. This may involve some personal frustration but it will be worth it.
Obtaining the Software
Instructions for installing the R language are here: http://cran.r-project.org/
Instructions for installing R Studio are here: http://www.rstudio.com/
Course Description
The course will cover:
- The major types of R variables: vectors (numerical, character, logical), matrices, data frames and lists.
- The important classes: numeric, character, list and changing between them
- Importing data from Excel
- Dealing with non-numeric instrument data: the “<10”s and “>1000”s.
- Manipulating your data: sub-setting, which, match, sort, unique, cut
- Simple statistical tests: mean, median, quantiles (normal ranges), t-tests, ANOVA, Wilcoxon.
- Data merges: matching rows between sets
- Exporting data to Excel-like format.
- Regressions: Ordinary Least Squares,Passing Bablok, Deming, weighted regressions.
- Non-linear regressions
- Looping: Doing things repeatedly
- Writing your own functions
- Making highly customized graphs: scatter plots, regression lines, histograms, box plots, qq-plots
- Putting it all together projects:
- Preparing method comparison regression and Bland Altman plots
- Preparing mass spectrometry data for upload to LIS.
EarlyBird Deadline |
Regular Deadline |
After |
|
| Student / Post-Doc (Trainee) | €84 | €100 | €135 |
| Academic / Non-Profit | €250 | $300 | €405 |
| Commercial / Industry | €420 | €505 | €670 |
Notice: Undefined index: days in /home/msacl0017/public_html/include/webpage_structure/include_subtab_content_nextgen.php(1236) : eval()'d code on line 220
Data Science 201 :: Going Further With R: Tackling Clinical Laboratory Data Manipulation and Modeling
| Level: | Intermediate | ||||||||||||||||||||
| Prereqs: |
| ||||||||||||||||||||
| Location: | Mozart 3 | ||||||||||||||||||||
| Instructor(s): | Patrick Mathias, MD, PhD & Shannon Haymond, PhD | ||||||||||||||||||||
| |||||||||||||||||||||
Patrick Mathias, MD, PhD, completed his undergraduate degree in Electrical Engineering at Duke University, followed by a master’s degree in Electrical and Computer Engineering from the University of Illinois at Urbana-Champaign. He then completed a MD degree and a PhD in Bioengineering from the University of Illinois, with a focus on nanophotonics and label-free biosensors. He completed residency training in Clinical Pathology as well as a Clinical Informatics fellowship at the University of Washington. He is currently the Associate Director of the Informatics division in the Department of Laboratory Medicine at the University of Washington. His clinical and research interests lie in improving electronic health record systems to improve ordering and interpretation of laboratory tests, developing infrastructure for novel analytical technologies in the clinical laboratory, and applying analytics to improve laboratory operations and clinical care at a population level. Co-Instructor: Shannon Haymond, PhD
Ann and Robert H. Lurie Children’s Hospital of Chicago, and My research interests are focused on the evaluation and utilization of novel biomarkers in pediatric diseases and combining systems-based approaches with patient reported measures of quality of life to better understand individual variation in treatment response. Clinically, I am interested in the development and validation of mass spectrometry (LC-MS/MS) assays and in the integration and predictive analysis of diverse data. | |||||||||||||||||||||
| |||||||||||||||||||||
Notice: Undefined variable: late_reg in /home/msacl0017/public_html/include/webpage_structure/include_subtab_content_nextgen.php(1236) : eval()'d code on line 444
Overview: Having completed your first steps into the wonderful world of data analysis with R, would you like to go further? You’ve learned the basics of R, so now it’s time to put that knowledge to work and tackle some interesting clinical applications. Along the way you will also be introduced to even more of capabilities of R and the tools developed by the amazing R community.
The course will be run over 16 hours and time will be split between lecture sessions, individual problem solving, and a highly interactive group-level data mining of real data sets (there may even be prizes). Like the introductory course, this class will maintain the “no student left behind policy”. Students will be given time to solve problems taken from real life laboratory work and to do some more advanced analysis on large scale data sets. All attendees will need to bring a laptop with the R language installed and R Studio interface installed. Students may use Windows, Mac OSX or Linux environments. Both R and R studio are free (as in “Free Beer”) and open-source.
Students should be prepared continue to expand their skill in programming – which, as you learned in the introductory course can be a little frustrating, but not as frustrating as not being able to get the computer to do what you want at all!
Obtaining the Software
Instructions for installing the R language are here: http://cran.r-project.org/
Instructions for installing R Studio are here: http://www.rstudio.com/
Course Description
The course will cover:
- Core concepts in reproducible research
- Introduction to R-Markdown for report generation
- Conceptual basis for keeping data “tidy”
- Using the Import -> Tidy -> Transform -> Visualize -> Model -> Communicate pipeline
- Data wrangling and manipulation operations such as filtering, grouping, summarizing
- Data exploration and visualization with the ggplot2 library
- Functional programming concepts for efficient iteration (purrr’s map functions)
- Introduction to machine learning workflows
- Prediction with linear regression and classification with logistic regression
EarlyBird Deadline |
Regular Deadline |
After |
|
| Student / Post-Doc (Trainee) | €84 | €100 | €135 |
| Academic / Non-Profit | €250 | $300 | €405 |
| Commercial / Industry | €420 | €505 | €670 |
Notice: Undefined index: days in /home/msacl0017/public_html/include/webpage_structure/include_subtab_content_nextgen.php(1236) : eval()'d code on line 220
Glyco-Proteomics 101 :: Unravelling The Sweetness Of Life: Clinical Glyco(Proteo)Mics By Mass Spectrometry
| Level: | Beginner to Intermediate | ||||||||||||||||||||
| Prereqs: | |||||||||||||||||||||
| Location: | Mozart 2 | ||||||||||||||||||||
| Instructor(s): | Noortje de Haan, PhD & Guinevere Lageveen-Kammeijer, PhD | ||||||||||||||||||||
| |||||||||||||||||||||
Noortje de Haan, PhD, is conducting her post-doctoral research at the Center for Proteomics and Metabolomics at the Leiden University Medical Center, The Netherlands. She obtained her MSc degree in Pharmaceutical Sciences at the VU University Amsterdam, The Netherlands, with a focus on biomarker research and clinical chemical analysis. She received her PhD at the Leiden University Medical Center, under the supervision of prof. Manfred Wuhrer, on the development and application of various mass spectrometry-based methods for the analysis of (antibody) glycosylation. Noortje’s enthusiasm for glycoproteomic-related research started early in her scientific career and this remains a key drive in her current work. She is interested in the development of mass spectrometric methods and data analysis protocols for addressing clinical research questions. Guinevere S.M. Lageveen-Kammeijer, PhD, received her PhD on exploring prostate-specific antigen (PSA), the well-known biomarker for prostate cancer, and its glycosylation by capillary electrophoresis and mass spectrometry. Currently, Guinevere performs her post-doctoral research at the Center for Proteomics and Metabolomics at the Leiden University Medical Center, in the group of prof. Manfred Wuhrer. She currently works on further expanding a mass spectrometry-based PSA glycosylation assay which she developed during her PhD. In addition, she explores the possibilities for the in-depth analysis of glycans and intact glycoproteins for biomarker discovery for other diseases as well as for the characterization of biopharmaceuticals. In 2017, Guinevere joined the organization committee of the Netherlands Area Biotech (NLab) Discussion group of CASSS. In 2019, she became a member of the scientific committee of the glycomics session, and a member of the early career committee, of MSACL EU. Her research interests are focused on bringing together researchers from the field of biomarker discovery with clinical laboratory professionals, ensuring a better translation of potential biomarkers to the clinic. Moreover, she is dedicated to convincing her fellow colleagues that glycosylation is an important subject and should not be neglected just because it is rather complicated. | |||||||||||||||||||||
| |||||||||||||||||||||
Notice: Undefined variable: late_reg in /home/msacl0017/public_html/include/webpage_structure/include_subtab_content_nextgen.php(1236) : eval()'d code on line 444
Overview: Did you ever encounter glycans, but you -kind of- neglected them as they seemed too complicated to characterize? Or did you just perform a glycan release to make the analysis of your protein a lot easier? Do you have no idea how to interpret your data when a glycan is present? Fear no more! We are here to provide you with the basics in the field of mass spectrometric glycomics and glycoproteomics.
The course will start with a historical overview on glycan research (i.e. how did glycans work their way up to being acknowledged as important study objects) and we will guide you through the maze of different nomenclatures. Moreover, although glycans are well known for their complexity, we will reveal to you the “rules of glycan structures” based on known biosynthetic pathways. This will be followed by an in-depth discussion on glyco(proteo)mic mass spectrometric technologies and workflows. In addition, different sample preparation steps will be covered. We will close-up with a session about glycomic biomarker discovery, as it can hardly be considered a coincidence that, for example, more than 80% of the currently used cancer biomarkers are glycosylated.
The course will run over two days and time will be split between lectures and workshops (e.g. how do you recognize a glycan in a mass spectrum and how do you assign it). While not everything can be covered within these two days we will ensure that you will know your “glyco-basics” in the end. Moreover, participants are encouraged to submit any specific glyco-questions they have prior to the course and we will try to discuss them during the course.
Brief outline of the course:
- Historical overview of glycosylation research
- Glycans from a chemical perspective
- The biosynthetic pathway of N-glycans, O-glycan and glycosphingolipids
- Biological function of glycans
- Historical overview of glycoanalytics
- How to analyze and interpret your glycan, glycopeptide and/or glycoprotein with mass spectrometry
- Glycomic biomarker discovery - Finding the sweet spot of the disease
EarlyBird Deadline |
Regular Deadline |
After |
|
| Student / Post-Doc (Trainee) | €84 | €100 | €135 |
| Academic / Non-Profit | €250 | $300 | €405 |
| Commercial / Industry | €420 | €505 | €670 |
Notice: Undefined index: days in /home/msacl0017/public_html/include/webpage_structure/include_subtab_content_nextgen.php(1236) : eval()'d code on line 220
LC-MSMS 101 :: Getting Started with Quantitative LC-MS/MS in the Diagnostic Laboratory
| Level: | Beginner | ||||||||||||||||||||
| Prereqs: | None. | ||||||||||||||||||||
| Location: | Paracelsus Hall | ||||||||||||||||||||
| Instructor(s): | Laura Owen, PhD, Michael Wright, Coral Munday & Grace van der Gugten | ||||||||||||||||||||
| |||||||||||||||||||||
Laura Owen is a Consultant Clinical Scientist at Salford Royal Hospital, where mass spectrometry assays for a variety of small molecule and toxicology services are offered nationally. Prior to working at Salford Laura worked at Wythenshawe Hospital where she has experience of quantitative method development on both LC-MS/MS and exact mass equipment. Laura is a honorary senior lecturer at the University of Manchester, where she lectures on chromatography, mass spectrometry and the adrenal cortex on a national clinical science MSc programme. Laura has also acted as chair of the Association for Clinical Biochemistry’s special interest group for mass spectrometry. Mike Wright is the Scientific Director at the Drug Development Solutions division of LGC, in Fordham, Cambridgeshire, UK. His team work on the development of assays for the monitoring of drugs and biomarkers, as drug development tools, in a wide variety of matrices. Prior to working at LGC, Mike worked in clinical diagnostics developing LC-MS/MS services for health trusts in the UK and Australia. Mike teaches on LC-MS/MS applications, considerations and troubleshooting and has contributed to a number of online training programs including the AACC’s Introductory Liquid Chromatography Mass Spectrometry certificate. Grace Van Der Gugten is LC-MS/MS Applications Development Specialist at St Paul’s Hospital in Vancouver BC. She is passionate about developing the most user friendly and streamlined LC-MS/MS assays as possible for routine use in the Special Chemistry Mass Spec Lab. She loves troubleshooting, especially when the cause of problem has been discovered and the issue solved!
| |||||||||||||||||||||
| |||||||||||||||||||||
Notice: Undefined variable: late_reg in /home/msacl0017/public_html/include/webpage_structure/include_subtab_content_nextgen.php(1236) : eval()'d code on line 444
Overview: Is your laboratory under pressure to purchase an LC-tandem MS or have the instruments arrived and your team are beginning on their mass spectrometry journey? This short course is designed for attendees implementing quantitative LC-tandem MS for patient testing who have laboratory medicine experience but no mass spectrometry training - clinical bench analysts, supervisors, R&D scientists, and laboratory directors. After starting out with a basic chemistry refresher, theoretical concepts necessary for a robust implementation of clinical mass spectrometry will be presented, however, the main focus of the course will be on practical recommendations for:
- Starting with MSMS, LC and sample extraction parameters
- Choosing internal standards, solvents, and water, making reagents and calibrators
- How to fine tune sample preparation, LC and MSMS parameters to achieve the desired assay performance
- Pre-validation stress testing and method validation
- Preventative maintenance and troubleshooting
- Maintaining quality once your new LC-MS/MS assay has gone live
- LC-MS/MS system purchasing
- site preparation and installation (dos and don’ts)
- establishing data analysis & review criteria and an interface to the LIS
The course will be a steady mixture of lectures, tutorials and problem solving sessions with our goal being to present just enough theory so you can report high quality results, while opening a window to the depth and complexity of clinical mass spectrometry such that your appetite is whetted to learn more. Previous exposure to the principles of clinical method validation, either theoretical or practical, is assumed. A glossary of common LC-MSMS terms/acronyms, and diagrams delineating basic LC and MSMS instrument components and functions will be emailed to attendees a week prior to the beginning of the course. This material will also be addressed at the beginning of the course, but the initial learning curve can be steep and review prior to the course will be beneficial if you have absolutely no previous exposure with LC-MSMS.
EarlyBird Deadline |
Regular Deadline |
After |
|
| Student / Post-Doc (Trainee) | €84 | €100 | €135 |
| Academic / Non-Profit | €250 | $300 | €405 |
| Commercial / Industry | €420 | €505 | €670 |
Notice: Undefined index: days in /home/msacl0017/public_html/include/webpage_structure/include_subtab_content_nextgen.php(1236) : eval()'d code on line 220
LC-MSMS 301 :: Development and Validation of Quantitative LC-MS/MS Assays for Use in Clinical Diagnostics
| Level: | Advanced | ||||||||||||||||||||
| Prereqs: | The target audience should have extensive familiarity with LC-MS/MS systems. | ||||||||||||||||||||
| Location: | Mozart 4-5 | ||||||||||||||||||||
| Instructor(s): | Brian Rappold & Chris Shuford, PhD | ||||||||||||||||||||
| |||||||||||||||||||||
Brian Rappold is the Director of Mass Spectrometry at LabCorp. He is a renowned expert in the field of method development and validation of mass spectrometry assays for clinical diagnostic use, teaching courses on this subject at numerous scientific conferences. His extensive knowledge and clinical perspective has granted him several opportunities to present his research internationally on the topics of hydrophilic interaction liquid chromatography for bio-analysis, multiplexed mass spectrometric detection of amino acidopathies and the origins and solutions of ion suppression in electrospray ionization. He currently serves as the chair of Clinical Chemistry for the American Society for Mass Spectrometry (ASMS) and has served on the Metabolomics and Small Molecule Analysis/Toxicology Scientific Committee for The Association for Mass Spectrometry: Applications to the Clinical Laboratory (MSACL). His research interests include the realization of open-access mass spectrometric systems and antibody-capture/mass spectrometry applications to diagnostic medicine. Chris Shuford, Ph.D., is Technical Director for research and development at LabCorp’s Center for Esoteric Testing in Burlington, North Carolina. Chris received his B.S. in Chemistry & Physics at Longwood University and obtained his Ph.D. in Bioanalytical Chemistry from North Carolina State University under the tutelage of Professor David Muddiman, where his research focused on applications of nano-flow chromatography (<500 nL/min) for multiplexed peptide quantification using protein cleavage coupled with isotope dilution mass spectrometry (PC-IDMS). In 2012, Chris joined LabCorp’s research and development team where his efforts have focused on development of high-flow chromatographic methods (>1 mL/min) for multiplexed and single protein assays for clinical application. | |||||||||||||||||||||
| |||||||||||||||||||||
Notice: Undefined variable: late_reg in /home/msacl0017/public_html/include/webpage_structure/include_subtab_content_nextgen.php(1236) : eval()'d code on line 444
Overview: This 16-hour course will briefly introduce the key aspects of the LC-MS/MS experimental workflow and then focus on processes and experimental designs for assay development and analytical validation of assays to be employed within clinical diagnostics.
The first day will describe method development in detail, including how-to guides for initial optimization of mass spectrometry systems, chromatographic development and sample preparation schemes. Techniques and technologies for streamlining analytical performance will also be described. Transitional experiments from development to validation will be discussed in detail to stress test methodologies prior to analytical validation.
Day two will cover all details pertinent in validation of LC-MS/MS analytical workflows. Experimental designs for all aspects of validation, putative acceptance criteria and analytical solutions will be shown. Key validation criteria of selectivity, carry-over, matrix effect, accuracy, precision, linearity, stability and inter-assay correlation will be described using multiple case studies.
EarlyBird Deadline |
Regular Deadline |
After |
|
| Student / Post-Doc (Trainee) | €84 | €100 | €135 |
| Academic / Non-Profit | €250 | $300 | €405 |
| Commercial / Industry | €420 | €505 | €670 |
Notice: Undefined index: days in /home/msacl0017/public_html/include/webpage_structure/include_subtab_content_nextgen.php(1236) : eval()'d code on line 220
Lipidomics 101 :: Mass Spectrometry-based Lipidomics and Clinical Applications
| Level: | Beginner - Intermediate | ||||||||||||||||||||
| Prereqs: | Interest in LC-MS, clinical sciences, laboratory medicine, etc. AND a joint interest in lipids. | ||||||||||||||||||||
| Location: | Papageno Hall | ||||||||||||||||||||
| Instructor(s): | Anne K Bendt, PhD, Amaury Cazenave-Gassiot, PhD, Michael Chen, MD | ||||||||||||||||||||
| |||||||||||||||||||||
With a MSc in Marine Biotechnology (Greifswald University, Germany) and a PhD in Biochemistry (Cologne University, Germany) in close collaboration with industry, Anne Bendt has always been working on topics which are translatable into applications outside of the laboratory. Driven by her fascination for infectious diseases, she joined the National University of Singapore (NUS) in 2004 to develop lipidomics tools for tuberculosis studies. She is now a Principal Investigator at the Life Sciences Institute, NUS, focussing on translation of mass spec technologies into clinical applications, primarily for lipids and small molecules. Additionally, she is serving as the Associate Director of the Singapore Lipidomics Incubator (SLING) being in charge of operations and commercialization. Anne is passionate about training and education and has made substantial contributions to SLING’s various workshops and the highly successful ‘ic lipid’ training course. Research Assistant Prof. Cazenave-Gassiot is an early-career researcher and an expert in mass spectrometry-based lipidomics. He graduated with a PhD in analytical chemistry at the University of Southampton (UK), under the supervision of Dr John Langley, specialising in supercritical fluid chromatography and mass spectrometry. His interest in lipids started while a postdoc in the team of Professor Anthony Postle, still in Southampton. A member of SLING since 2009, his research centres on separation sciences, mass spectrometry, and their applications to life sciences, especially lipid biochemistry. He has developed chromatographic and mass spectrometric methods for the identification and quantification of lipids in diverse biological systems. This has included successful local and international collaborations. Michael Chen did his undergraduate degree in Biochemistry, and subsequently obtained his MD from McGill University. After residency in Laboratory Medicine at McGill, he completed his Master’s Degree (McGill University and University of Victoria) as part of the Clinician Investigator Program in Experimental Medicine with a focus on Clinical Proteomics. He also received fellowship training in clinical lipidology. Michael is also a Medical co-Director at the Victoria Lipid Clinic. Dr Chen’s research interests focus on clinical and translational mass spectrometry. His current medical practice and research emphasizes on the following domains: (1) management of inherited dyslipidemia (2) biomarker verification and validation using mass spectrometry-based technique (3) implementation of mass spectrometry assays in clinical laboratory (4) standardization and utilization of remnant biospecimens for biobanking. | |||||||||||||||||||||
| |||||||||||||||||||||
Notice: Undefined variable: late_reg in /home/msacl0017/public_html/include/webpage_structure/include_subtab_content_nextgen.php(1236) : eval()'d code on line 444
Overview: This 8-hr course is meant to (1) create awareness for the importance and therefore potential value of lipid testing beyond cholesterol and triglycerides for future clinical applications. We will (2) then outline currently available technologies and their respective opportunities and challenges, and (3) discuss candidate molecules in the context of two case studies. (4) Finally, this short course is designed to be engaging thus will require some active participation via online live-feedback.
1) Looking beyond cholesterol and TAG:
- Gain an understanding of the universe of lipids, how they are intricately linked to biology and their implications in health and diseases
- Potential of blood based lipid testing
- Identify physiologically relevant candidate lipids for adoption by the clinical community, for future studies towards establishing clinical utility
2) Current lipidomics R&D workflows:
- Path of translation from R&D laboratory-style methods towards robust and quantitative assays with appropriate turnaround times
- Pre-analytics (sampling requirements, plasma vs serum, storage, etc.)
- Analytics (i.e. batches, internal standards, lipid extractions, direct infusion vs LC-MS and LC-MS/MS, quality assurance)
- Post-analytics (raw data processing, lipid annotations, quality control, quantification),
- Ongoing harmonization efforts
3) Case studies of markers that have advanced to clinical settings:
- Examples of markers that have already been implemented in a clinical setting in larger scale studies, to illustrate the feasibility and technical tractability, the clinical impact of lipid testing and its value on overall healthcare
4) Outreach and Engagement between the analytical scientist specialized in mass spectrometry of lipids, the clinician researcher and laboratory medicine as the end user are key to the development of impactful / useful lipidomics in clinical applications.
EarlyBird Deadline |
Regular Deadline |
After |
|
| Student / Post-Doc (Trainee) | €60 | €72 | €90 |
| Academic / Non-Profit | €180 | $216 | €270 |
| Commercial / Industry | €300 | €360 | €450 |
Notice: Undefined index: days in /home/msacl0017/public_html/include/webpage_structure/include_subtab_content_nextgen.php(1236) : eval()'d code on line 220
Metabolomics 202 :: Metabolomics: Approaches, Applications and Challenges
| Level: | Beginner to Intermediate | ||||||||||||||||||||
| Prereqs: | LC/MS hands on experience. | ||||||||||||||||||||
| Location: | Doppler Hall | ||||||||||||||||||||
| Instructor(s): | Julijana Ivanisevic, PhD & Elizabeth Want, PhD | ||||||||||||||||||||
| |||||||||||||||||||||
Julijana Ivanisevic, PhD is the head of the Metabolomics Research Platform (Senior Lecturer) at the Faculty of Biology and Medicine, University of Lausanne. Her research expertise is in mass spectrometry-based metabolomics applications to biomedical research (brain metabolism, ageing, cancer metabolism), evolutionary biology and ecology. Her postdoctoral research was at the Center for Metabolomics at The Scripps Research Institute in La Jolla, California, USA. Elizabeth Want, PhD is a Senior Lecturer in Molecular Spectroscopy in the Department of Surgery and Cancer at Imperial College, London and the Director of the Imperial International Phenome Training Centre. She joined Imperial College in 2006 after working as a postdoctoral researcher at the Scripps Research Institute in La Jolla, CA. Her research at Imperial College involves the development, optimisation and application of LC-MS methodologies for the analysis of biological samples, largely in the context of metabolic phenotyping. She applies these methods to biomedical research areas including toxicology, cardiovascular disease, neonatal disease and development, and neurological diseases. | |||||||||||||||||||||
| |||||||||||||||||||||
Notice: Undefined variable: late_reg in /home/msacl0017/public_html/include/webpage_structure/include_subtab_content_nextgen.php(1236) : eval()'d code on line 444
Overview:
Metabolome: Downstream of Genome
- Central Dogma of Molecular Biology – From Genotype to Metabotype
- Historical Perspective of Metabolomics
- Technological platforms (NMR, GC/MS, LC/MS) and Applications
- OmicChallenge – Metabolite Diversity
Approaches in Metabolomics
- Targeted versus Untargeted (work on the front end vs. work on the back end, instrumentation, etc.)
- Experimental Design and Sample Preparation (depending on the approach)
- Choice of Analytical Platform (depending on the approach and metabolites of interest)
- OmicChallenge – « Big » Data Reduction
Data (Pre)Processing in Untargeted Experiments
- Step by step from peak picking to peak grouping and annotation
- Open-access platforms
- Hands-on XCMS Online
- Omic Challenge – Metabolite Identification
Statistical Analysis, Metabolite Identification and Mapping onto Pathways
- Univariateversus Multivariate Statistics (SIMCA, MetaboAnalyst)
- Metabolite Matching against Metabolite Databases (METLIN, HMDB, LIPIDMAPS, MassBank)
- Metabolite Set Enrichement Analysis and Network Modeling (MetaboAnalyst, KEGG, BioCyc, Mummichog, Ingenuity)
- Omic Challenge – Integration with other Omic Technologies in a Biologically Relevant Context
EarlyBird Deadline |
Regular Deadline |
After |
|
| Student / Post-Doc (Trainee) | €84 | €100 | €126 |
| Academic / Non-Profit | €250 | $300 | €380 |
| Commercial / Industry | €420 | €505 | €630 |
Notice: Undefined index: days in /home/msacl0017/public_html/include/webpage_structure/include_subtab_content_nextgen.php(1236) : eval()'d code on line 220
Proteomic Microbiology 201 :: Bottom-Up and Top-Down Proteomic Approaches for Bacterial Identification and Characterization, a Focus on MALDI-TOF and Advanced Technologies
| Level: | Beginner - Intermediate | ||||||||||||||||||||
| Prereqs: | None. | ||||||||||||||||||||
| Location: | Trapp Zimmer | ||||||||||||||||||||
| Instructor(s): | Jean Armengaud, PhD, Stefan Zimmermann, MD | ||||||||||||||||||||
| |||||||||||||||||||||
Jean Armengaud, PhD is best known for his work on proteogenomics of bacteria and the characterization of pathogens and radiotolerant organisms. He directs a mass spectrometry research unit located near Avignon in France that is dedicated to proteomics-based identification and quantitation of pathogens and environmentally relevant bacteria. Stefan Zimmermann, MDis head of the division bacteriology at the department of infectious disease (University Hospital Heidelberg). He uses mass spectrometry for identification of bacteria and fungi in the routine workflow of the lab for up to ten years. He is interested in MS applications beyond identification, like antibiotic susceptibility testing and subtyping of pathogens. | |||||||||||||||||||||
| |||||||||||||||||||||
Notice: Undefined variable: late_reg in /home/msacl0017/public_html/include/webpage_structure/include_subtab_content_nextgen.php(1236) : eval()'d code on line 444
Overview: This course will present an overview of bottom-up and top-down techniques for microbial identification using mass spectrometry-based technologies as well as their use in determination of microbial characteristics such as antibiotic resistance profiles. Topics to be discussed include:
- Comparison of bottom-up versus top-down proteomic approaches,
- Why top down proteomics is well-suited for microbial identification and characterization,
- Discrimination of closely related strains by top-down proteomics approaches.,
- Collection and interpretation of MALDI-TOF data,
- Proteogenomics as a means for improving annotations based on genomic sequence analysis and its use in identification of key protein markers in MALDI-TOF spectra.
- The concept of proteoforms as a means of categorizing the PTM states of proteins.
- The application of these techniques and technologies to antibiotic resistance determination,
- Novel methodologies that are currently emerging for the analysis of difficult samples, such as mixtures of pathogens and spores present within complex matrices.
- Recent advances for pathogen quantitation by tandem mass spectrometry.
- Use of MALDI-TOF for identification of viruses, molds and parasites.
EarlyBird Deadline |
Regular Deadline |
After |
|
| Student / Post-Doc (Trainee) | €60 | €72 | €90 |
| Academic / Non-Profit | €180 | $216 | €270 |
| Commercial / Industry | €300 | €360 | €450 |